Overview
Traditional Enzyme Directed Evolution
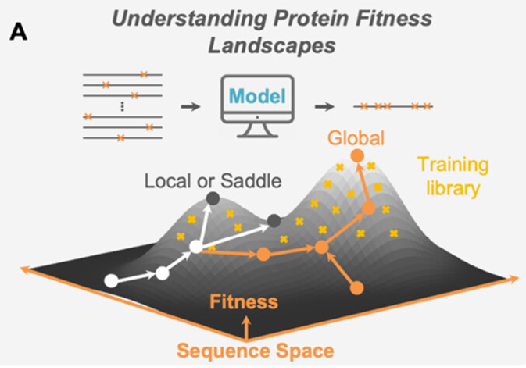
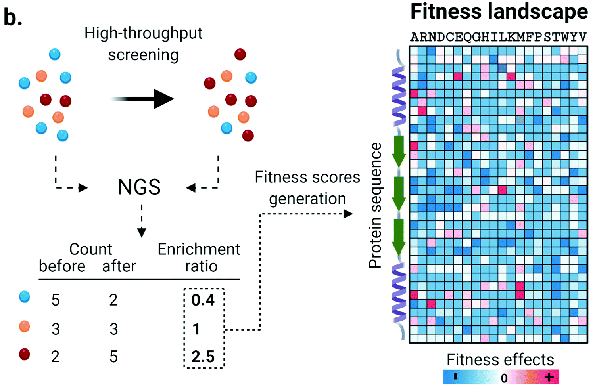
Obtaining mutant enzymes with improved performance through either completely random mutagenesis or random mutagenesis at specific sites combined with high-throughput screening typically requires multiple rounds of screening. Besides the immense workload of screening, a significant problem with this approach is that, given 20 different conventional amino acids, the total number of possible mutants for a protein with a typical length of 300 amino acids is 20300, far exceeding our screening capabilities. Even if we choose to randomly alter 10 sites, using the NNK codon to construct a mutant library, the number of variables still reaches 1.1 x 1015, which is still beyond our screening capabilities. Obviously, only a tiny fraction of the possible mutations are screened through conventional directed evolution, thus the probability of finding significant performance improvements is typically low.
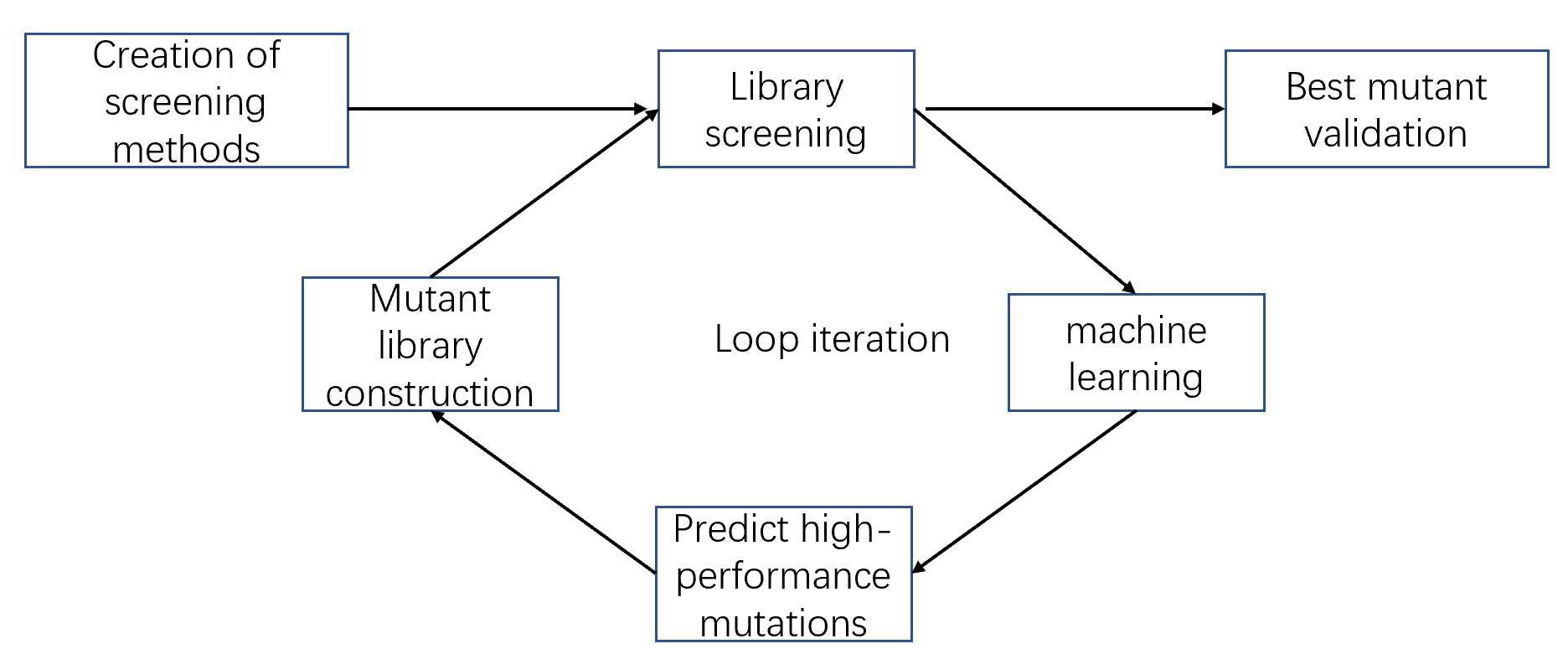
Machine learning-assisted enzyme engineering
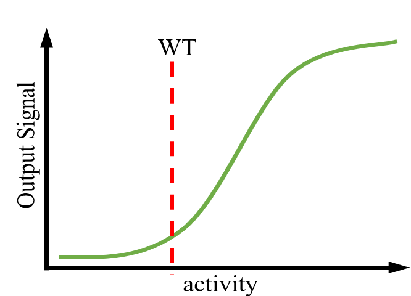
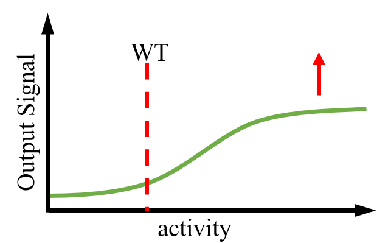
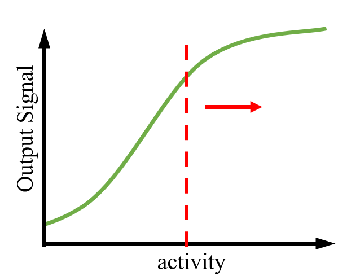
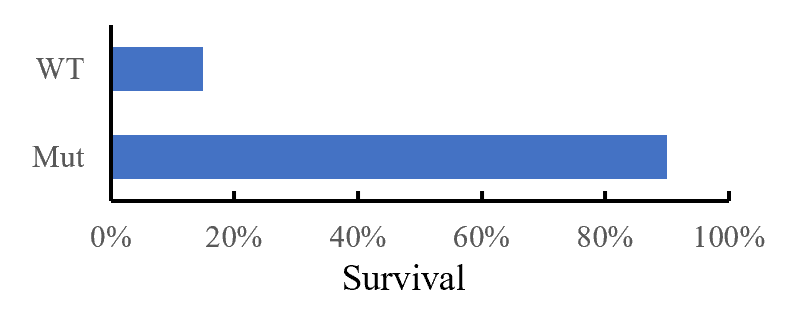
A controllable amount of mutant activity data is used in machine learning to predict highly active mutants, and only a smaller number of predicted sequences can be screened to obtain significantly improved mutants (DOI: 10.1038/s41592-021-01100- y). It greatly reduces the screening workload and time and improves the efficiency of enzyme engineering and design.